"Ordinary gene sequencing" should be referred to as "conventional DNA sequencing," which is a technique based on the Sanger method, also known as the dideoxy method. Today, automatic sequencing using instruments like the ABI 3730xl is widely adopted, and it can typically produce reads ranging from 600 to 800 base pairs.
The term "high-throughput sequencing" is actually a relative concept. In the year 2000, sequencing performed on machines such as the 3700 and MegaBace was considered high-throughput, compared to manual methods or gel-based sequencing. However, after 2005, the term began to refer specifically to second-generation sequencing technologies, such as 454, Solexa (later renamed Illumina), and SOLiD. These platforms offer a dramatic improvement in throughput—often tens of thousands to hundreds of millions of times more efficient than first-generation sequencers like the 3730.
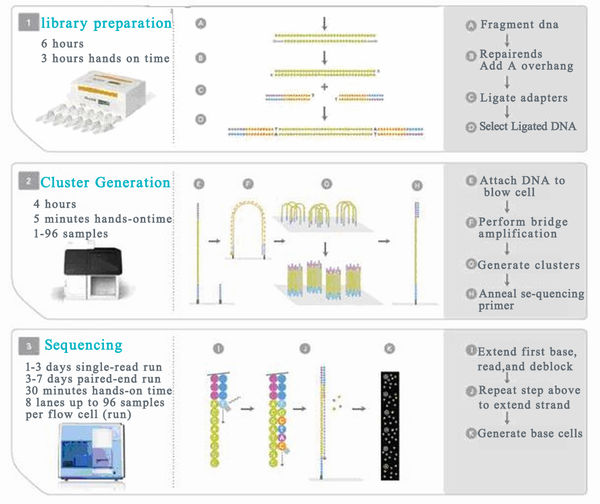
The main features of Next-Generation Sequencing (NGS) include:
1. High throughput: A single run can generate between 500 Mb to 600 Gb of data.
2. Short read lengths: For example, 454 produces about 400–500 bp, while Illumina generates 100–250 bp, and SOLiD offers 75–100 bp.
3. Very low cost per unit of data: The cost of sequencing has significantly dropped, making large-scale projects more feasible. As a result, bioinformatics analysis has become even more critical in interpreting the data.
What is high-throughput gene sequencing?
It is a process that builds upon the dideoxy sequencing method introduced by Sanger et al. in 1977. To sequence both ends of each clone insert, each plasmid template plate requires two 384-well cycle sequencing plates. The sequencing reaction uses Big Dye Terminator chemistry version 3.1 from Applied Biosystems, along with standard M13 or commonly used forward and reverse primers. This process is automated using a Biomek FX pipetting station, where a robotic arm aliquots the template and mixes it with the reaction solution. The solution includes dideoxynucleotides, fluorescently labeled nucleotides, Taq DNA polymerase, primers, and buffer. Barcodes on the templates and plates ensure accurate tracking during transfer.
A linear amplification step is then carried out on thermal cyclers like the MJ Research Tetrads or 9700 Thermal Cycler. The product is efficiently precipitated at room temperature using isopropanol and stored at 4°C. If the sequencing instrument is functioning properly, a sample film is automatically generated for each reaction plate after scanning its barcode. The plate is then transferred to an ABIPrism 3700 or Applied Biosystems 3730xi DNA Analyzer for electrophoresis. Modern systems allow up to 8 runs per day on the 3700 and 12 on the 3730xl, with a setup time under one hour.
High-throughput sequencing equipment that processes a large number of samples in parallel usually relies on automated laboratory information management and sample tracking systems (LIMS). At institutions like TIGR, these systems manage everything from early sequencing to end-of-sequence tracking, including library construction. Data is stored in a Sybase relational database, allowing users to trace back from annotated genes to original sequencing files.
These systems also support client/server applications for sample management, data entry, library handling, and sequence processing. Over time, they have evolved to incorporate new lab techniques, instruments, and software, resulting in a mature and stable platform. Integrated tools include automated vector removal, identification and masking of repetitive elements, detection of contaminated clones, and tracking of template information.
Throughout the sequencing process, the quality of templates and sequences can be monitored daily through an intuitive interface. This ensures that any issues are quickly identified and resolved. Quality control teams work closely with production teams, inspecting reagents, monitoring template quality, detecting deviations, and ensuring data accuracy. They also develop standard operating procedures to maintain consistency and technical precision.
The significance of high-throughput sequencing cannot be overstated. It marks a major milestone in genomics research, drastically reducing the cost of nucleic acid sequencing compared to earlier methods. For instance, human genome sequencing at the end of the last century cost around $3 billion, while second-generation sequencing has brought this down to the era of the "10,000 genome." Such cost efficiency enables genome projects for more species, helping us decode the genetic code of a wider range of organisms. It also makes large-scale whole-genome resequencing possible for species that already have their genomes sequenced.
High Capacity Pvc Pe Pipe Production Line,Precision Pvc Pe Pipe Production Line,Affordable Pvc Pe Pipe Production Line,Customizable Pvc Pe Pipe Production Line
Zhejiang IET Intelligent Equipment Manufacturing Co.,Ltd , https://www.ietmachinery.com